EPIGENETICS
| Staff(s) | Associate Professor. Diana Buzas |
|---|---|
| Field of research | Epigenetics |
| Research topics | Epigenetic mechanisms for cellular memory mediated by DNA demethylation and Polycomb |
| Keywords | DNA demethylation, DNA glycosylase, Iron-sulfur cluster, Polycomb and Tritorax group proteins, Polycomb recruiting elements, noncoding RNAs, Flowering locus C |
| URL | https://sites.google.com/view/dianabuzaslab |
| TEL | 029-853-6685 |
| buzas.mihaela.ka@u.tsukuba.ac.jp |
Research introduction

Feel free to contact me to talk about our research projects, life as a foreigner in Japan, scientific career, financial support etc if you are considering studying/working at University of Tsukuba. I am looking forward to hear from you!
Requirements for iron-sulfur clusters in DNA demethylation in Arabidopsis thaliana
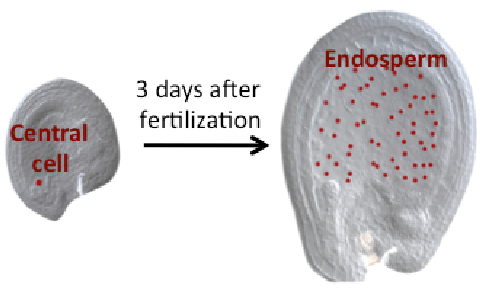
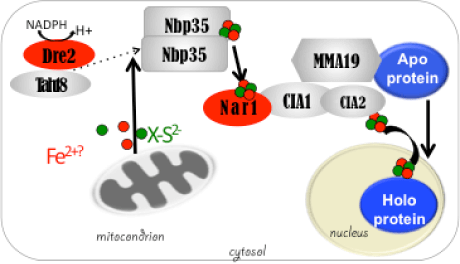
DME excises 5mC via the base excision repair pathway (BER) and is dependent on co-factors termed Iron-Sulfur (Fe-S) clusters. Fe-S clusters are ubiquitous in all cellular compartments where dedicated Fe-S biogenesis machineries exist. Nuclear proteins, including DME, are expected to obtain Fe-S clusters via the cytosolic iron sulfur cluster biogenesis pathway (CIA). Our work implicated the two CIA proteins DRE2 (Buzas 2014) and AtNar1 (Nakamura et al 2013) in this process. DRE2 has activity towards DNA demethylation in the same spatiotemporal window with DME at least six DME gene targets. Like DME, DRE2 binds Fe-S clusters and is present in the nucleus. However, because most other CIA pathway proteins do not appear to control 5mC removal in the central cell (Buzas 2014), it is enigmatic how do DME and DRE2 obtain their Fe-S clusters.
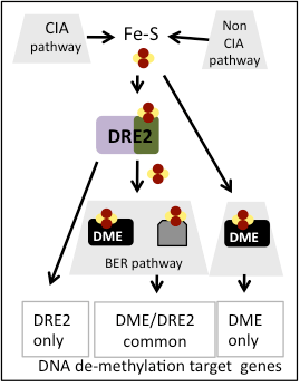
Working model
To explain the apparent paradox on Fe-S cluster role in 5mC removal, it is possible that genes targeted by this process have different sensitivity for Fe-S clusters because they are regulated by different mechanisms, i.e., DRE2 only-dependent, DME-only dependent, DRE and DME–dependant. Moreover, these genes may have different contribution to detectable phenotypes (some DME target genes do not have a phenotype when mutated). Furthermore, it is possible that DRE2 and/or DME obtain Fe-S clusters via a non-CIA pathway.
Ongoing projects
- Mitochondria control of nuclear genome integrity and DNA methylation
- Genetic interaction between components of the CIA and BER pathways
- The role of AtDRE2 in stress responses in Arabidopsis thaliana
References
-
Buzas D, Nakamura M, Kinoshita T. Epigenetic role for the conserved Fe-S cluster biogenesis protein Dre2 in Arabidopsis thaliana. PNAS. 2014 111(37); 13565-70.
http://www.yokohama-cu.ac.jp/univ/pr/press/pdf/140902_res.pdf
https://www.youtube.com/watch?v=BAUa78vfID0 - Nakamura M, Buzas DM, Kato A, Fujita M, Kurata N, Kinoshita T. The role of Arabidopsis thaliana NAR1, a cytosolic iron-sulfur cluster assembly component, in gametophytic gene expression and oxidative stress responses in vegetative tissue. New Phytologist 2013 Sep;199(4):925-35
Collaborators on this project
| Prof Testu Kinoshita |
Yokohama City University, Kihara Institute of Biological Research http://tetsu-kinoshita.jp |
|---|---|
| Prof Tsutomu Kawasaki |
Kinki University http://www.kindai.ac.jp/meikan/482-kawasaki-tsutomu.html |
| Assist Prof Hiroyuki Tsuji |
Yokohama City University, Kihara Institute of Biological Research |
Polycomb/Tritorax function underlying cellular memory of winter in annual and perennial crucifers
Many plants flower in spring. It is the cold of winter which controls flowering in some species, but plants must not flower when it is cold but rather after cold, in favorable spring conditions. In one plant family, Brassicacea, flowering is controlled by a repressor of flowering gene, Flowering Locus C (FLC), which is negatively regulated by prolonged cold (this process is called vernalization). The regulation of FLC by vernalization is linked with the type of life history a species adopts. When FLC expression remains repressed even after the end of winter (i.e., they have a permanent memory of winter) plants flower and terminate life in the same year, (annual species). When FLC expression after winter is progressively de-repressed (i.e., the memory of winter is transient) plants live on for another cycle (perennial species).
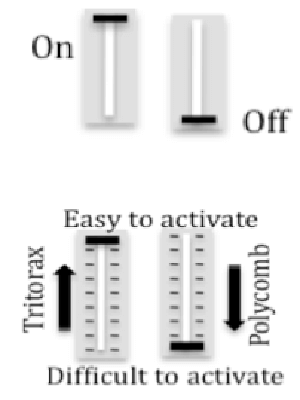
How is FLC expression controlled? Genetic mechanisms for gene regulation are generally either activating (on) or repressing (off). In contrast, Polycomb and Trithorax epigenetic mechanisms alter chromatin to create a repertoire of alternative transcription states, depending on the relative cellular levels of Polycomb, Trithorax and transcriptional regulators. The repressed chromatin state elevates the threshold for a minimum activator concentration sufficient for triggering transcription of a target gene, and the gene thereby becomes less sensitive to activation. In contrast, when in the active chromatin state, a gene depends to a lesser extend on the concentration of transcriptional activators and it is less sensitive to the presence of repressors. When cells within a cell population adopt different transcriptional states a quantitative response may be generated.
FLC expression is controlled epigenetically by the antagonizing action of Polycomb and Trithorax, but how are annual and perennial memories of winter determined is still unknown.
Working model
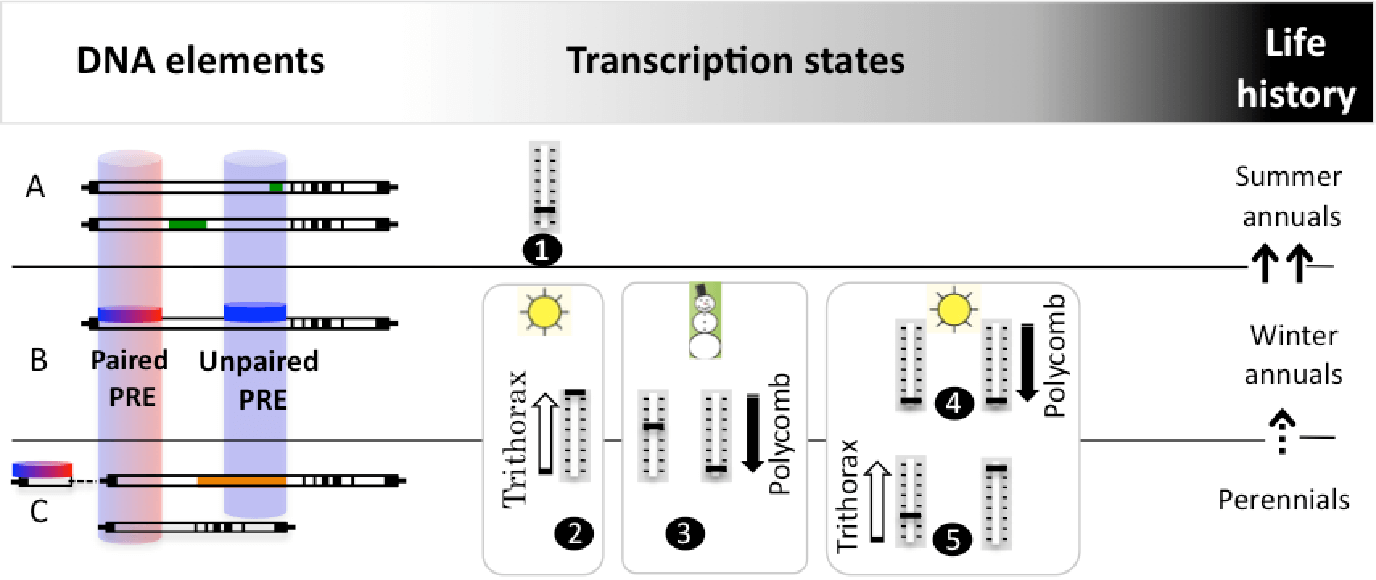
The vernalization response relies on alternative FLC transcription states, established at PREs (Polycomb Response Elements) located mainly within non-coding regions of the first long intron (white rectang are introns; exons and untranslated regions are represented in black). Our studies on winter annuals, which complete life cycle in two seasons (B) demonstrated that FLC has two PREs ( Buzas et al 2011, Buzas et al 2012). The promoter-proximal PRE associates with a TRE (Thritorax Response Element) (I term this a paired PRE, red and blue disc/cylinder). The active state before cold (B,2) and the quantitative repressed state in the cold (B,3; left, early cold, right, late cold) are controlled from this proximal PRE. The permanent repressed state after cold (B,4) is attributed to a distal PRE (blue cylinder), capable of recruiting exclusively Polycomb, which I term an unpaired PRE.
Rapidly cycling summer annuals (A) evolved many times from winter annuals (arrows) by loosing the active transcriptional state (A,1), via two routes. At the DNA level, modifications of mainly the first intron lead to weak FLC alleles. These sequences are known, but have not been investigated for PRE activity. In two of them, it appears that the distance between the paired and unpaired PRE had increased (top and bottom sequences each have an insertion – green rectangles).
In perennials (C), information on PREs is also lacking, but intriguingly, from the limited sequence information available, it appears that the unpaired PRE may be absent (top sequence, Arabis alpina: distal half of first intron is not conserved (shaded in orange) and the proximal sequence is partially duplicated; bottom sequence, Brassica oleracea: the first intron is shortened to the proximal half).
I predict that the life history is determined by the distribution of the paired and unpaired PREs at FLC that dictate its alternative expression states.
Ongoing projects
- Defining promoter and polycomb recruiting cis acting elements at FLC chromatin in Arabidopsis halleri gemifera
- Identification and functional analysis of noncoding RNAs from FLC locus in Arabidopsis halleri gemifera
- Function of Polycomb recruiting elements in Arabidopsis
References
- Deng W, Buzas DM, Ying H, Robertson M, Taylor J, Peacock WJ, Dennis ES, Helliwell C. Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genomics. 2013 Aug 30;14:593.
- Buzas DM, Tamada Y, Kurata T FLC: A Hidden Polycomb Response Element Shows Up in Silence. Plant Cell Physiology 2012 May;53(5):785-93.
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES. Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One. 2011;6(6):e21513.
- Buzas DM, Robertson M, Finnegan EJ, Helliwell CA. Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. Plant Journal 2011 (65): 872-881.
- Jean Finnegan E, Bond DM, Buzas DM, Goodrich J, Helliwell CA, Tamada Y, et al. Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant Journal 2011 Feb;65(3):382-91.
- Luo, M, Luo, MZ, Buzas D, Finnegan J, Helliwell, C, Dennis et al. A.UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 2008 180, 229-236.
Collaborators on this project
| Prof Hiroshi Kudoh |
Kyoto University, Center for Ecological Research http://www.ecology.kyoto-u.ac.jp/~kudoh/en/index.html |
|---|---|
| Assoc Prof Akiko Satake |
Kyushu University |
| Assist Prof Yosuke Tamada |
National Institute for Basic Biology |


